|
We recently welcomed two new members of the lab: Christina who is visiting us from Aalborg University to develop an environment specific database using long-read amplicon sequencing, and Thomas as a master student who will explore metagenomic datasets for plastid genomes.
But we are also getting ready to see Mahwash graduate with PhD, which will be a very special moment for us (first PhD student of the lab) but also sad because it means that she will soon leave. We are happy to announce the publication of two papers, now available at:
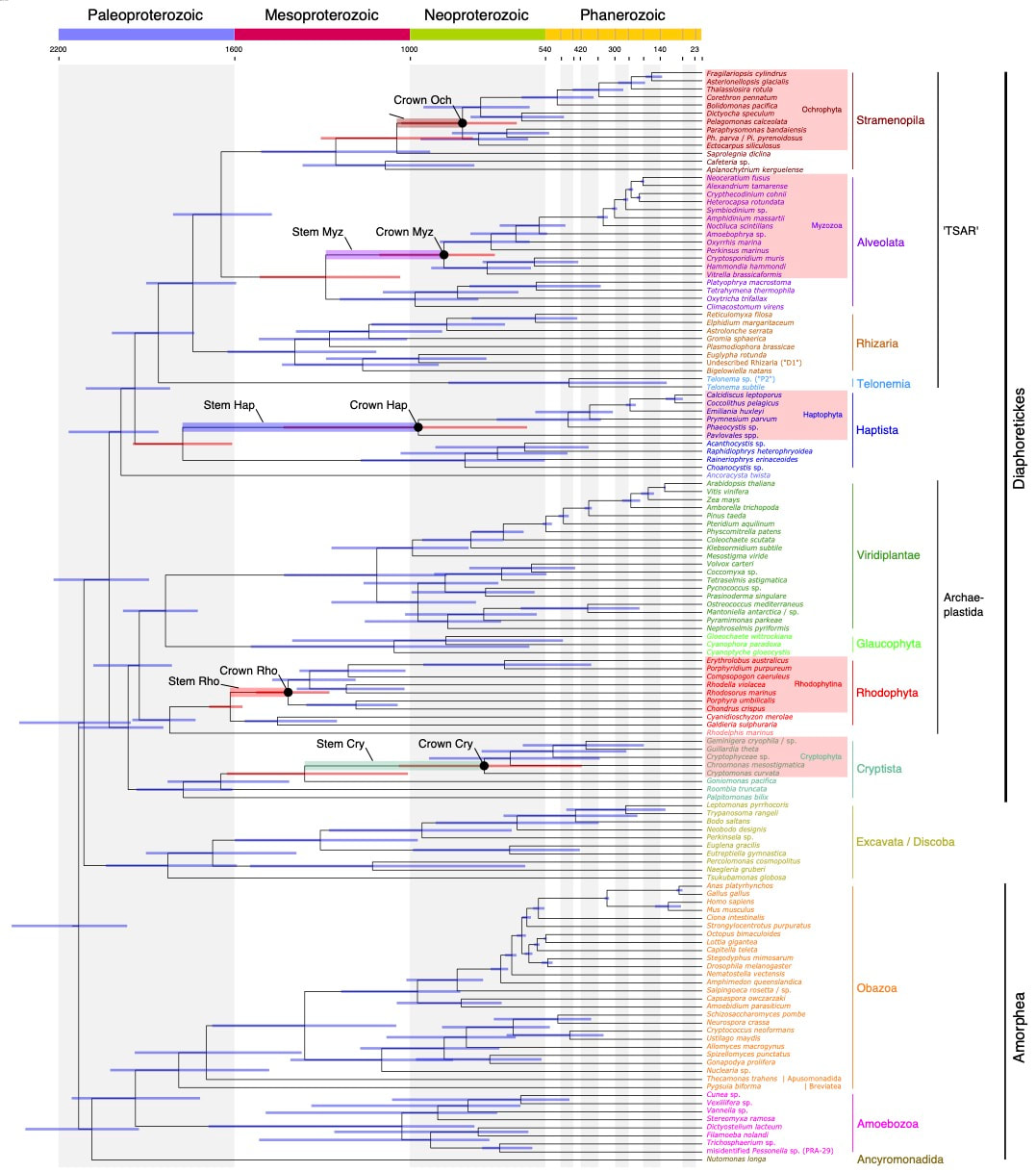
Iker Irisarri, Jürgen F H Strassert, Fabien Burki, Phylogenomic Insights into the Origin of Primary Plastids, Systematic Biology, 2021;, syab036, https://doi.org/10.1093/sysbio/syab036 Zlatogursky, V.V., Shɨshkin, Y., Drachko, D. and Burki, F. (2021), The Long-time Orphan Protist Meringosphaera mediterranea Lohmann, 1902 [1903] is a Centrohelid Heliozoan. Journal of Eukaryotic Microbiology. Accepted Author Manuscript e12860, https://doi.org/10.1111/jeu.12860 Extremely happy to present our new study with a phylogenomic timescale for the broad evolution of eukaryotes. Link to the OA paper: https://www.nature.com/articles/s41467-021-22044-z
A bunch of us decided to go grilling in the forest :-) It also coincided with the end of Charlie's project with us :-( coincide
Heidi Ottesen has joined the lab for a few months, she will do a project work with us on assembling a bunch of parasite genomes. Welcome Heidi!
A book chapter in Methods in Molecular Biology on how to use PREQUAL, a tool to remove non-homologous residues in phylogenomic datasets: https://link.springer.com/protocol/10.1007/978-1-0716-1036-7_10
We 'recently' welcomed three new members in the lab: Megan Sorensen (post-doc, in collaboration with the lab of main PI Rachel Foster at Stockholm University), Charlie Biwer (MSc student), and Miguel Mendez Sandin (postdoc). We're very excited to have them joined the lab!
Two good news in the lab recently, Fabien was appointed Associate Professor with tenure (Senior Lecturer) and received his docent in biodiversity informatics.
|