Our paper on the discovery of leptophytes, a novel major algal group, is now out in Nature Communications. Compared to the initial preprint, which was exclusively based on plastid MAGs (ptMAGs), this published version presents a leptophyte mitochondrial MAG (mtMAG).

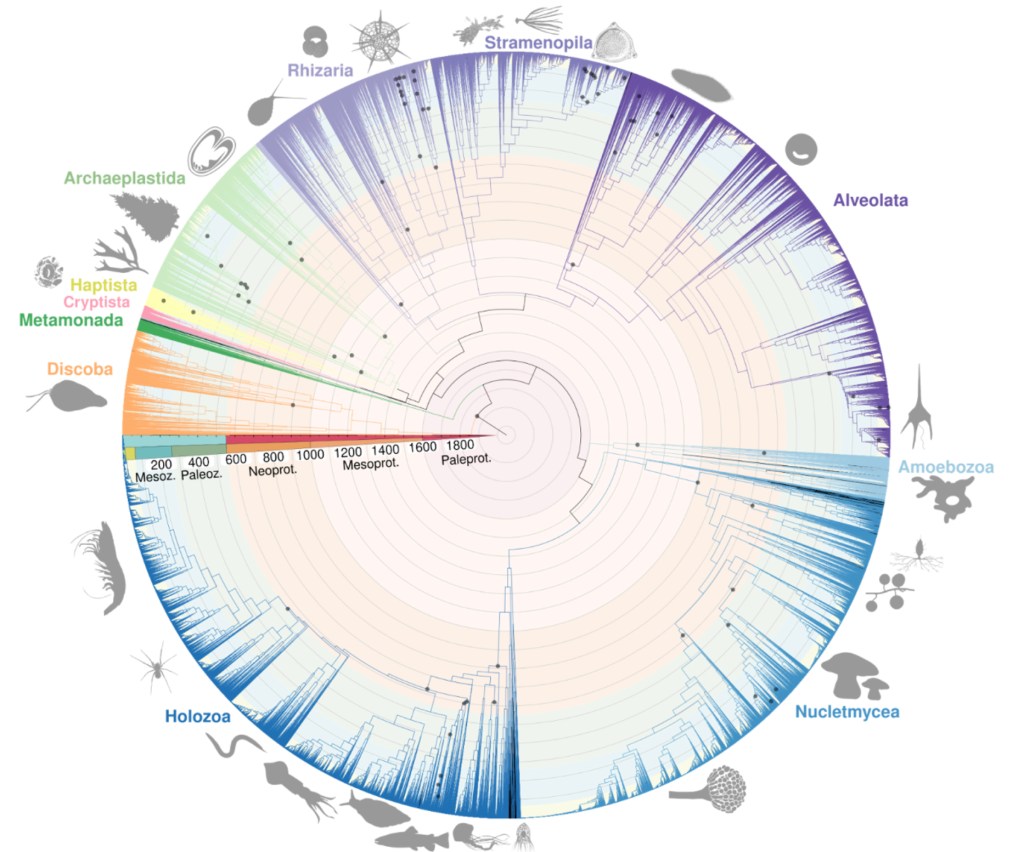
We also deposited a preprint entitled “Environmental phylogenetics supports a steady diversification of crown eukaryotes starting from the mid Proterozoic”. We present a unique phylogenetic dataset of over 75K OTUs, integrating information from the large but often discarded environmental diversity using long‐read metabarcoding data to studying the diversification dynamics of all major eukaryotic groups
from the Proterozoic until today.